SNARE family members associate into stable complexes that direct membrane fusion events. The SNARE mediated process of vesicle Ginsenoside-F4 exocytosis is not only critical for phagocytosis but is also required for the release of inflammatory cytokines from macrophages and microglia. SNARE complexes prime vesicles for fusion with the plasma membrane and are composed of proteins from each of three separate families: a SNAP, a Syntaxin, and a VAMP. SNARE complexes are formed by cells in anticipation of membrane fusion events and upon stimulation, the complexes localize to the points of vesicle contact, facilitate membrane fusion, and then dissociate into their individual components that are recycled to form new complexes. Numerous studies in other labs have shown that a-syn may affect SNARE complex assembly either through direct interaction with members of the SNARE family, or by sequestering agents, such as arachadonic acid, which promote SNARE assembly and activity. We have recently developed a novel transgenic genomic mouse model in which wild type or the E46K mutant form of human asyn was expressed from a human bacterial artificial chromosome. This model is similar to that created by Kou et al. however, we focused on wild type and the E46K mutated form of a-syn rather than the A30P and A53T mutations described previously. The BAC construct contains the a-syn promoter and other potential regulatory regions including upstream, and 15 kb downstream sequences. Because a-syn expression is driven from its own promoter, not from a highly active exogenous promoter, this in vivo system may provide insight into human disease, because in humans duplication or triplication of the gene is sufficient to lead to early onset PD. In addition, 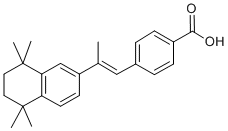 the potential impact of elevated asyn levels on multiple Sipeimine cellular and tissue systems can be assessed. In the work described here, we used these mice to examine the role of a-syn in innate immune cell function. We were particularly interested in macrophages and microglia because of their involvement in PD pathology and their intensive reliance on membrane dynamics for their central roles in cytokine release and phagocytic function. Our data demonstrates that increased expression of the a-syn protein has an inhibitory effect on the release of inflammatory cytokines, vesicular fusion, and phagocytic processes. Defective phagocytosis occurred in brain microglia and peripheral macrophages, either isolated and assayed in culture, or assayed in situ. Fibroblasts and monocytes isolated from sporadic PD patients and a patient carrying a triplication of the a-syn gene exhibited elevated a-syn levels, which correlated with defective phagocytic function. We also expand upon the molecular details regarding asyn interaction with membrane trafficking components, and demonstrate active recruitment of a-syn to a site of stimulated membrane fusion during phagocytosis. Our data further defines the role that a-syn plays in membrane vesicle dynamics and extends these findings into systemic processes beyond neuronal function, which may provide a valuable peripheral biomarker of elevated a-syn levels and dysfunctional vesicle dynamics. Our data suggest that there may be a previously unappreciated involvement of innate immune cell function in synucleinopathies, and that cells of the innate immune system provide an excellent, tightly regulated cellular model in which to study the effects of increased a-syn levels on vesicle function. Our data demonstrate that a-syn is expressed in innate immune cells and that the level of a-syn expression can regulate immune cell function.
the potential impact of elevated asyn levels on multiple Sipeimine cellular and tissue systems can be assessed. In the work described here, we used these mice to examine the role of a-syn in innate immune cell function. We were particularly interested in macrophages and microglia because of their involvement in PD pathology and their intensive reliance on membrane dynamics for their central roles in cytokine release and phagocytic function. Our data demonstrates that increased expression of the a-syn protein has an inhibitory effect on the release of inflammatory cytokines, vesicular fusion, and phagocytic processes. Defective phagocytosis occurred in brain microglia and peripheral macrophages, either isolated and assayed in culture, or assayed in situ. Fibroblasts and monocytes isolated from sporadic PD patients and a patient carrying a triplication of the a-syn gene exhibited elevated a-syn levels, which correlated with defective phagocytic function. We also expand upon the molecular details regarding asyn interaction with membrane trafficking components, and demonstrate active recruitment of a-syn to a site of stimulated membrane fusion during phagocytosis. Our data further defines the role that a-syn plays in membrane vesicle dynamics and extends these findings into systemic processes beyond neuronal function, which may provide a valuable peripheral biomarker of elevated a-syn levels and dysfunctional vesicle dynamics. Our data suggest that there may be a previously unappreciated involvement of innate immune cell function in synucleinopathies, and that cells of the innate immune system provide an excellent, tightly regulated cellular model in which to study the effects of increased a-syn levels on vesicle function. Our data demonstrate that a-syn is expressed in innate immune cells and that the level of a-syn expression can regulate immune cell function.
Month: May 2019
Annotation is historically developed for human consumption rather than for computational interpretation
Essentially, this means textual annotations are mostly made up of free-text English. Although textual Pimozide annotation studies are limited, several explore ways to model propagation of structural annotation errors. Structural annotation sits between nucleotide sequences and textual annotations; it identifies genomic elements, 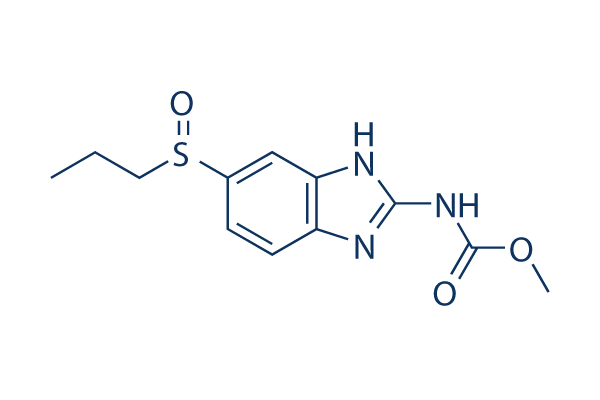 such as open reading frames, for a given sequence. This is Danshensu similar to textual annotation, in that structural annotation often makes use of sequence data and can be manually or automatically curated. These studies highlight a number of reasons for structural annotation errors, such as mis-identification of homology, omissions or typographical mistakes, concluding that annotation accuracy declines as the database size increases. Further studies attempt to actually estimate the error rates in structural annotation. These include an estimated error rate of between 28% and 30% in GOSeqLite and between 33% and 43% in UniProtKB/Swiss-Prot. Therefore, it is highly plausible that these errors will affect textual annotation, as acknowledged by Gilks et al.. We hypothesise that sentence reuse is prominent within textual annotations and a lack of formal provenance has led to inaccuracies in the annotation space. Within this paper we aim: to quantify sentence reuse; to investigate patterns of reuse and provenance, through a novel visualisation technique; and to investigate whether we can use patterns of propagation to identify erroneous textual annotations, inconsistent textual annotations or textual annotations with low confidence. A typical sentence within our work is one which contains a group of words and is terminated with a full stop. However, there are a number of exceptions to this basic rule, such as abbreviations, which are especially commonplace in the biomedical domain. The vast majority of these are handled correctly by LingPipe. We have shown that sentence reuse in UniProtKB is both common and increasing. Therefore, given the scale of this data, we explore the usage of visualisation. By visualising sentence reuse across entries and over time, we may be able to better understand annotation propagation and infer provenance. From this, it is possible that patterns demonstrating interesting traits in the underlying data may emerge and be identified. Therefore, we wish to explore how we can visualise this data and ask: how can we clearly represent the flow of annotation through the database? A number of approaches to visualising large datasets were considered. One such approach is to model the relationship between sentences and entries as a graph. Using a tool, such as Cytoscape, we can easily model sentences occurring within entries. However, our experience with this approach suggests that it is troublesome to model change over time and manual intervention is often required to ensure nodes are organised in a correct and meaningful manner. Other similar approaches, such as Sankey diagrams, were not utilised as we cannot determine the exact source and flow of an annotation between each individual entry. One approach which produces a visualisation similar to our requirements is the history flow tool. This tool was developed to allow visualisation of relationships between multiple versions of a wiki. Therefore, it aims to clearly depict the change in sentences, and their order, in a document over time with the ability to attribute each change to a given author. The authors demonstrated this visualisation with an exploratory analysis of Wikipedia, revealing complex patterns of cooperation and conflict between Wikipedia authors. However, using the history flow tool to visualise the flow of individual sentences in UniProtKB is not ideal.
such as open reading frames, for a given sequence. This is Danshensu similar to textual annotation, in that structural annotation often makes use of sequence data and can be manually or automatically curated. These studies highlight a number of reasons for structural annotation errors, such as mis-identification of homology, omissions or typographical mistakes, concluding that annotation accuracy declines as the database size increases. Further studies attempt to actually estimate the error rates in structural annotation. These include an estimated error rate of between 28% and 30% in GOSeqLite and between 33% and 43% in UniProtKB/Swiss-Prot. Therefore, it is highly plausible that these errors will affect textual annotation, as acknowledged by Gilks et al.. We hypothesise that sentence reuse is prominent within textual annotations and a lack of formal provenance has led to inaccuracies in the annotation space. Within this paper we aim: to quantify sentence reuse; to investigate patterns of reuse and provenance, through a novel visualisation technique; and to investigate whether we can use patterns of propagation to identify erroneous textual annotations, inconsistent textual annotations or textual annotations with low confidence. A typical sentence within our work is one which contains a group of words and is terminated with a full stop. However, there are a number of exceptions to this basic rule, such as abbreviations, which are especially commonplace in the biomedical domain. The vast majority of these are handled correctly by LingPipe. We have shown that sentence reuse in UniProtKB is both common and increasing. Therefore, given the scale of this data, we explore the usage of visualisation. By visualising sentence reuse across entries and over time, we may be able to better understand annotation propagation and infer provenance. From this, it is possible that patterns demonstrating interesting traits in the underlying data may emerge and be identified. Therefore, we wish to explore how we can visualise this data and ask: how can we clearly represent the flow of annotation through the database? A number of approaches to visualising large datasets were considered. One such approach is to model the relationship between sentences and entries as a graph. Using a tool, such as Cytoscape, we can easily model sentences occurring within entries. However, our experience with this approach suggests that it is troublesome to model change over time and manual intervention is often required to ensure nodes are organised in a correct and meaningful manner. Other similar approaches, such as Sankey diagrams, were not utilised as we cannot determine the exact source and flow of an annotation between each individual entry. One approach which produces a visualisation similar to our requirements is the history flow tool. This tool was developed to allow visualisation of relationships between multiple versions of a wiki. Therefore, it aims to clearly depict the change in sentences, and their order, in a document over time with the ability to attribute each change to a given author. The authors demonstrated this visualisation with an exploratory analysis of Wikipedia, revealing complex patterns of cooperation and conflict between Wikipedia authors. However, using the history flow tool to visualise the flow of individual sentences in UniProtKB is not ideal.
For interaction between the tissue source and the effect of PE or gestational age on transcription values
Galectins are highly expressed at the maternal-fetal interface, and regarded as multifunctional regulators of fundamental cellular processes due to their capacity to modulate functions such as cell-extracellular matrix interactions, proliferation, adhesion, and invasion. Galectin 3 is expressed in placental cell columns, but not in invasive extravillous trophoblasts. The negative correlation between expression and trophoblast invasiveness is in accordance with our finding of decreased galectin 3 expression over gestation. Galectin 8 is expressed by decidual cells, villous and extravillous trophoblasts, but its role is less clearly understood. The IPA analyses of the 22 PE-associated transcripts demonstrated an over-representation of genes associated with metabolic disease. PE share several metabolic abnormalities with cardiovascular diseases and diabetes, and having a PE complicated pregnancy is associated with increased risk of type 2 diabetes later in life. This agrees with pregnancy acting as a stress factor which could reveal a pre-existing 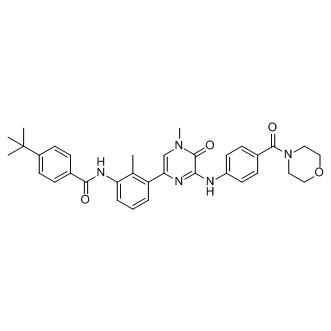 disposition to later life metabolic disease. IPA analysis of the 92 transcripts that were associated with gestational age revealed an over-representation of genes involved in cell assembly and organisation, tissue development, cellular movement, tissue morphology, and connective tissue development and function. These findings are in agreement with known biological processes taking place at the maternal-fetal interface during pregnancy, such as trophoblast proliferation, differentiation, invasion and extracellular matrix remodelling. It is important to consider that the data used in our study was produced on two different microarray platforms, and that tissue sampling procedures differed between the three sub-studies included in our dataset. In dataset #1 and #2, basal plate biopsies were used for transcriptional analyses, whereas in dataset #3, decidual tissue was collected by vacuum suction of the entire placental bed. These differences may pose a potential bias, as gene expression has been shown to differ depending on tissue sampling site. Inter- and intra-platform reproducibility has been shown to be good in terms of detecting differentially expressed genes. However, the reproducibility of absolute Acetylcorynoline transcription levels is poor, and pre-processing is required before comparisons of transcription values across platforms can be made. To deal with these limitations, and be able to make both inter- and intraplatform comparisons of transcription values, we performed inverse normal transformation. After this transformation, the distribution of transcription values is assumed equal for all probes, independent of study, sampling method and platform. However, PE-status, gestational age and tissue sampling method differed between studies, and consequently we had to include this as a factor Ginsenoside-Ro giving a separate level for each study in the linear regression model. In our analyses, we exclusively searched for observations that agreed between datasets, and the limitations mentioned above will rather reduce the power of identifying differentially expressed genes than introduce false positive results. Combined with the fact that our pooled dataset only targets 14,678 genes, and that we are using a very stringent threshold for significance, our results likely represent only a small fraction of the total number of transcripts that are influenced by either PE or gestational age. Had all samples been collected by the same method and analysed on the same arrays, it is likely that a much larger number of differentially expressed transcripts would be identified.
disposition to later life metabolic disease. IPA analysis of the 92 transcripts that were associated with gestational age revealed an over-representation of genes involved in cell assembly and organisation, tissue development, cellular movement, tissue morphology, and connective tissue development and function. These findings are in agreement with known biological processes taking place at the maternal-fetal interface during pregnancy, such as trophoblast proliferation, differentiation, invasion and extracellular matrix remodelling. It is important to consider that the data used in our study was produced on two different microarray platforms, and that tissue sampling procedures differed between the three sub-studies included in our dataset. In dataset #1 and #2, basal plate biopsies were used for transcriptional analyses, whereas in dataset #3, decidual tissue was collected by vacuum suction of the entire placental bed. These differences may pose a potential bias, as gene expression has been shown to differ depending on tissue sampling site. Inter- and intra-platform reproducibility has been shown to be good in terms of detecting differentially expressed genes. However, the reproducibility of absolute Acetylcorynoline transcription levels is poor, and pre-processing is required before comparisons of transcription values across platforms can be made. To deal with these limitations, and be able to make both inter- and intraplatform comparisons of transcription values, we performed inverse normal transformation. After this transformation, the distribution of transcription values is assumed equal for all probes, independent of study, sampling method and platform. However, PE-status, gestational age and tissue sampling method differed between studies, and consequently we had to include this as a factor Ginsenoside-Ro giving a separate level for each study in the linear regression model. In our analyses, we exclusively searched for observations that agreed between datasets, and the limitations mentioned above will rather reduce the power of identifying differentially expressed genes than introduce false positive results. Combined with the fact that our pooled dataset only targets 14,678 genes, and that we are using a very stringent threshold for significance, our results likely represent only a small fraction of the total number of transcripts that are influenced by either PE or gestational age. Had all samples been collected by the same method and analysed on the same arrays, it is likely that a much larger number of differentially expressed transcripts would be identified.
As the molecular mechanisms behind impaired trophoblast invasion are preferentially reflected at the maternal-fetal interface
They no longer require attachments for their survival and thus achieve anoikis resistance. Twist plays a critical role in breast cancer metastasis. Furthermore, high Twist expression in D-Pantothenic acid sodium infiltrative EC affects patient survival, though the mechanism of EMT in EC Mepiroxol remains unclear. Results from our study support high levels of Twist in EC, which are significantly associated with TrkB expression. Furthermore, we show that levels of Twist expression are modulated by TrkB and are required for TrkB-induced EMT transformation and tumorigenesis in vitro. Notably, in anchorage-independent culture, we found that both TrkB and Twist were expressed in survival cell spheroids, and many EMT-related genes were differentially expressed compared to adherent cells, which are hallmarks of the oncogenic EMT being required for anoikis resistance. Peritoneal dissemination of EC cells is a multistep process consisting of invasion into the serosa from the uterus, detachment from the primary site, movement into the peritoneal cavity, attachment to the distant peritoneum, invasion into the subperitoneal space, and proliferation. From our in vivo experiments, we discovered that the TrkB knockdown significantly inhibited the establishment of intraperitoneal disseminated tumors. A constitutive “cadherin switch” and apoptosis shift were also detected in paraffin-embedded sections of mouse tumors, which are consistent with the in vitro results. Mechanistically, Trks are receptor tyrosine kinases activated by neurotrophins and other growth factors. TrkA, TrkB and TrkC are the preferred receptors for the neurotrophins NGF, BDNF and neurotrophin-3, respectively. When stimulated by BDNF ligand, TrkB induces the activation of various downstream signaling pathways including Akt, Src, or MAPK resulting in cell proliferation, and apoptosis resistance in models of human cancer. In the present study, we found that both BDNF and TrkB are over-expressed in human endometrial carcinoma specimens. In vitro experiments verified that BDNF enhances cell proliferation and survival. In addition, the migratory and invasive properties of EC are mediated by a BDNF-TrkB signaling cascade in Ishikawa and RL95-2 cells. After the discovery of the involvement of the BDNF/TrkB cascade in cancer biology, it is appreciated that the inhibition of TrkB activity might be beneficial in a clinical oncology setting. Previous studies have demonstrated 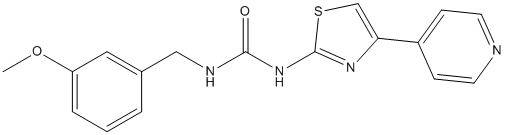 a key role for BDNF-TrkB signaling in modulating the response to cytotoxic agents, and modulation of TrkB expression enhanced the sensitivity of cells to cis-Diamminedichloroplatinum Cisplatin in head and neck squamous cell carcinoma. Recently, It was reported Trk inhibitor K252a inhibited cell growth and induced apoptotic cell death in uterine leiomyosarcoma.Therefore, further studies are needed to explore the development of small-molecule compounds that act as TrkB antagonists, or monoclonal antibodies against either BDNF or TrkB, as promising novel therapies for the treatment of endometrial carcinoma. Our findings provide new insights into biological mechanisms of EC and establish TrkB as a potential target for future therapies for this disease. The aetiology of PE is not completely understood, but it is generally considered that disturbed interactions between the invading trophoblasts and maternal cells causing defective trophoblast invasion are important pathophysiological events. The subsequent impaired spiral artery remodelling and reduced placental perfusion is proposed to create oxidative stress and a release of inflammatory factors into the maternal circulation, causing overt PE. Gene expression analyses may provide further insight in mechanisms of disease and function as preventive, predictive or therapeutic measures.
a key role for BDNF-TrkB signaling in modulating the response to cytotoxic agents, and modulation of TrkB expression enhanced the sensitivity of cells to cis-Diamminedichloroplatinum Cisplatin in head and neck squamous cell carcinoma. Recently, It was reported Trk inhibitor K252a inhibited cell growth and induced apoptotic cell death in uterine leiomyosarcoma.Therefore, further studies are needed to explore the development of small-molecule compounds that act as TrkB antagonists, or monoclonal antibodies against either BDNF or TrkB, as promising novel therapies for the treatment of endometrial carcinoma. Our findings provide new insights into biological mechanisms of EC and establish TrkB as a potential target for future therapies for this disease. The aetiology of PE is not completely understood, but it is generally considered that disturbed interactions between the invading trophoblasts and maternal cells causing defective trophoblast invasion are important pathophysiological events. The subsequent impaired spiral artery remodelling and reduced placental perfusion is proposed to create oxidative stress and a release of inflammatory factors into the maternal circulation, causing overt PE. Gene expression analyses may provide further insight in mechanisms of disease and function as preventive, predictive or therapeutic measures.
In light of new knowledge a previous annotation may now be erroneous with respect to current knowledge
Given that annotations propagate, any updates to an original annotation should also be propagated. However, we identify over 8,000 sentences which may, or may have, incorrectly remained  in the database. While these first three patterns are of interest in regard to annotation quality, we next investigate whether we can use the missing root origin pattern as an indication for erroneous annotations. Current methods for detecting textual annotation provenance and tracking its propagation are somewhat limited. Within this paper we have presented a technique that allows annotation provenance and propagation to be identified and visualised by using sentences. Further, we have provided an analysis of sentence reuse levels and identified a number of annotation patterns that provide an indication as to an annotations quality and correctness. The cornerstone of this work was dependent upon sentence reuse in UniProtKB. Our analysis shows that reuse is heavily prevalent for both Swiss-Prot and TrEMBL. This is because of the curation process employed by UniProt, which consists of six key stages, one of which involves Gambogic-acid identifying similar entries and standardising annotations between these entries. If two entries from the same gene and species are identified then they are merged. Therefore, sentences are effectively copied between entries as a matter of protocol. This process can see sections, or sometimes whole annotations, from one entry being copied to other entries without change. Whilst the levels of reuse are generally increasing overtime, we interestingly note a slight decline in sentence reuse for later versions of Swiss-Prot. Although this decline coincides with the change of the UniProtKB release cycle, it appears to be related to a change in annotation policy for Swiss-Prot. After 2010 only sequences with experimental annotation were added to Swiss-Prot; previously automatically annotated orthologue sequences from complete genomes were often included in Swiss-Prot. In the face of ever increasing raw biological data, this reuse is not unexpected. Whilst manual curation is often regarded as the ��gold standard’, it is a significant bottleneck. For example, in the FlyBase database it can take between two and four months for an article to be manually curated with consideration recently being given to incorporating sections of automated processing into the curation process. It was for this same reason that UniProtKB introduced TrEMBL in 1996. Whilst reuse is understandably higher within automated methods, it is inevitably going to remain commonplace throughout both automated and manual databases while the quantity of raw biological data being generated continues to increase. Indeed, sentence reuse is an important feature of annotation curation. In addition to the propagation of knowledge, it also allows annotations to become standardised and can be used to enforce levels of quality control. Whilst these results further highlight the importance of being able to identify the origin of an annotation, the analysis was only achievable given that UniProtKB make available all major historical versions of Swiss-Prot and TrEMBL. Users are typically only interested in the most recent and up-to-date biological data available, but this work highlights the added value and importance of being able to scour archival data; database features such as UniSave should be a requirement rather than a luxury. It was this archival data that allows provenance and propagation to be analysed, allowing the Cefetamet pivoxil HCl development of a visualization technique. These visualisations appear to be useful, as their usage allowed a number of propagation patterns to be identified.
in the database. While these first three patterns are of interest in regard to annotation quality, we next investigate whether we can use the missing root origin pattern as an indication for erroneous annotations. Current methods for detecting textual annotation provenance and tracking its propagation are somewhat limited. Within this paper we have presented a technique that allows annotation provenance and propagation to be identified and visualised by using sentences. Further, we have provided an analysis of sentence reuse levels and identified a number of annotation patterns that provide an indication as to an annotations quality and correctness. The cornerstone of this work was dependent upon sentence reuse in UniProtKB. Our analysis shows that reuse is heavily prevalent for both Swiss-Prot and TrEMBL. This is because of the curation process employed by UniProt, which consists of six key stages, one of which involves Gambogic-acid identifying similar entries and standardising annotations between these entries. If two entries from the same gene and species are identified then they are merged. Therefore, sentences are effectively copied between entries as a matter of protocol. This process can see sections, or sometimes whole annotations, from one entry being copied to other entries without change. Whilst the levels of reuse are generally increasing overtime, we interestingly note a slight decline in sentence reuse for later versions of Swiss-Prot. Although this decline coincides with the change of the UniProtKB release cycle, it appears to be related to a change in annotation policy for Swiss-Prot. After 2010 only sequences with experimental annotation were added to Swiss-Prot; previously automatically annotated orthologue sequences from complete genomes were often included in Swiss-Prot. In the face of ever increasing raw biological data, this reuse is not unexpected. Whilst manual curation is often regarded as the ��gold standard’, it is a significant bottleneck. For example, in the FlyBase database it can take between two and four months for an article to be manually curated with consideration recently being given to incorporating sections of automated processing into the curation process. It was for this same reason that UniProtKB introduced TrEMBL in 1996. Whilst reuse is understandably higher within automated methods, it is inevitably going to remain commonplace throughout both automated and manual databases while the quantity of raw biological data being generated continues to increase. Indeed, sentence reuse is an important feature of annotation curation. In addition to the propagation of knowledge, it also allows annotations to become standardised and can be used to enforce levels of quality control. Whilst these results further highlight the importance of being able to identify the origin of an annotation, the analysis was only achievable given that UniProtKB make available all major historical versions of Swiss-Prot and TrEMBL. Users are typically only interested in the most recent and up-to-date biological data available, but this work highlights the added value and importance of being able to scour archival data; database features such as UniSave should be a requirement rather than a luxury. It was this archival data that allows provenance and propagation to be analysed, allowing the Cefetamet pivoxil HCl development of a visualization technique. These visualisations appear to be useful, as their usage allowed a number of propagation patterns to be identified.