Importantly, erlotinib was able to effectively block ligand-induced EGFR INCB18424 phosphorylation in all cell lines tested, indicating that the ability of erlotinib to block EGFR activation was not impaired even after cells developed resistance to its growth inhibitory effects. To further investigate the relationship of p-AKT, p-ERK1/2 and Mig6 to the sensitivity of erlotinib, we again blotted the 26 cell line panel and plotted protein kinase inhibitors expression level against the IC50 of erlotinib. Our data showed that Mig6 expression was associated better with p-AKT than p-ERK1/2, which suggested that p-AKT pathway might be playing bigger role in regulating Mig6. Interestingly, introduction of Mig6 to H292 cells significantly increased resistance to erlotinib when concomitantly decreased basal EGFR phosporylation was seen. However, it did not affect sensitivity to erlotinib in SCC-S cells where EGFR phosporylation was not affected. Taken together, our data suggested that cellular dependence on EGFR, which can be predicted by basal Mig6/EGFR ratio, underlie the response of cancer cells to erlotinib rather than the absolute expression level of Mig6. This was further supported by our observation that Mig6/EGFR demonstrated a high degree of accuracy as the predictor of EGFR activity in a large panel of head and neck, bladder and lung cancer cell lines examined. In addition, in review of data from a published report, the relative expression of Mig6 and EGFR also correlates well with basal EGFR activity in a panel of breast cancer cells examined. To understand whether Mig6 knockdown in combination with p-AKT inhibition sensitize cells to erlotinib, we knocked down Mig6 and treated cells with AKT inhibitor. We found that AKT pathway inhibition could be detrimental to the resistant cells over the period of a few days. However, co-treatment with low dose of AKT inhibitor did sensitize cells to erlotinib in H1703 cells. To investigate whether our observations with tumor cell lines could be validated in tumor samples from patients, we analyzed directly xenografted low passage human tumors that have been shown to retain the key features of the original tumor, including drug sensitivity, and that accurately represent the heterogeneity of the disease. We obtained 4 human NSCLCs, and 18 pancreatic tumors that were directly xenografted into nude mice. No erlotinib-sensitizing mutations in EGFR were detected in any of these tumors. We initially tested the response of the 4 patient-derived lung xenografts to erlotinib. Among them, BML-5 showed a better response to erlotinib than the other 3 tumors. Analysis of Mig6 expression in tumor xenografts showed that BML-1 and BML-5 expressed less Mig6 than BML-7 and BML-11. In addition, BML-5 expressed higher total EGFR as well as higher basal EGFR phosphorylation than the other tumors. We next characterized and plotted erlotinib responsiveness of 18 directly xenografted pancreatic tumors. Tumor growth inhibition data are displayed with the most sensitive tumors on the far left  and the most resistant on the far right. Tumor characteristics, including KRAS mutation status as well as EGFR expression and phosphorylation levels, have been reported previously. No EGFR sensitizing-mutations were found in any of these tumors and there was no correlation of KRAS mutation with erlotinib response in pancreatic tumors. EGFR negative tumors tended to cluster on the right side of the map, indicating that they were more resistant to erlotinib. However, in EGFR-positive tumors we saw little association between erlotinib sensitivity and EGFR expression. Instead, we found that in these pancreatic tumors, as Mig6 expression increased, tumors exhibited a more erlotinib-resistant phenotype. For example, the erlotinib-resistant tumor PANC420 expressed markedly higher Mig6 than the erlotinib-sensitive tumor PANC410, even though they expressed comparable amounts of EGFR protein. In keeping with their Mig6 expression status, PANC410 displayed heavy EGFR phosphorylation whereas PANC420 harbored no detectable EGFR phosphorylation. Interestingly, in the 3 erlotinib-resistant pancreatic tumors studied that displayed significantly lower Mig6 expression, IHC labeling revealed that 2 of these 3 xenograft lines did not express EGFR. Studies have suggested a weak association between EGFR protein expression levels and responsiveness to EGFR TKIs. Although the erlotinib-sensitive tumors studied here generally displayed high EGFR levels, our data suggested that it was the activity of EGFR.
and the most resistant on the far right. Tumor characteristics, including KRAS mutation status as well as EGFR expression and phosphorylation levels, have been reported previously. No EGFR sensitizing-mutations were found in any of these tumors and there was no correlation of KRAS mutation with erlotinib response in pancreatic tumors. EGFR negative tumors tended to cluster on the right side of the map, indicating that they were more resistant to erlotinib. However, in EGFR-positive tumors we saw little association between erlotinib sensitivity and EGFR expression. Instead, we found that in these pancreatic tumors, as Mig6 expression increased, tumors exhibited a more erlotinib-resistant phenotype. For example, the erlotinib-resistant tumor PANC420 expressed markedly higher Mig6 than the erlotinib-sensitive tumor PANC410, even though they expressed comparable amounts of EGFR protein. In keeping with their Mig6 expression status, PANC410 displayed heavy EGFR phosphorylation whereas PANC420 harbored no detectable EGFR phosphorylation. Interestingly, in the 3 erlotinib-resistant pancreatic tumors studied that displayed significantly lower Mig6 expression, IHC labeling revealed that 2 of these 3 xenograft lines did not express EGFR. Studies have suggested a weak association between EGFR protein expression levels and responsiveness to EGFR TKIs. Although the erlotinib-sensitive tumors studied here generally displayed high EGFR levels, our data suggested that it was the activity of EGFR.
Month: July 2019
Differences between the members of this family has resulted in their classification as PAKs
The type I PAKs are functionally and structurally well-studied, and are directly activated by BEZ235 interaction with Rho-family small GTPases to function in growth factor signaling and regulation of morphogenic processes. In contrast, the type II PAKs bind the Rho-family small GTPases CDC42, RAC1 and RhoV, but are not directly activated by this interaction. Instead, 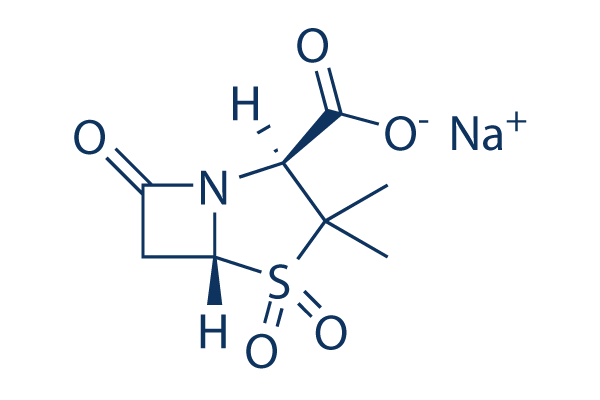 alternate mechanisms of activation and regulation have recently been discovered. The type II PAKs are important for signaling LY294002 cascades that regulate cell survival, neurite outgrowth and formation of filipodia. PAK6 is expressed in prostate, testis, thyroid, placenta and neural tissues and is found in both cytoplasmic and nuclear fractions of prostate cells. Androgen receptor is reported to be a downstream target of PAK6, and PAK6 can regulate gene transcription by androgen receptor via a GTPase-independent mechanism possibly related to control of its degradation by the MDM2 E3 ubiquitin ligase. Global deletion of Pak6 in mice results in increased weight and decreased aggression, possibly explained by its role in androgen receptor signaling. In addition, mice with combined deletion of Pak5 and Pak6 show deficits in locomotion, learning and memory not associated with single deletions of either gene, suggesting functional redundancy between the two PAKs. While neuronal substrates specific to PAK6 have not been identified, PACSIN1, an FBAR protein involved in synaptic vesicle recycling, is phosphorylated redundantly by PAK4, PAK5 and PAK6 in vivo PAK6 is overexpressed in prostate cancer, and its targeted inhibition could potentially decrease growth of prostate tumors or sensitize prostate cancer cells to radiotherapy. PAK6 has also been found to acquire somatic mutations in other solid tumors, including mutation of residue Pro52 to leucine in two independent melanomas. Consequently there is increasing interest in obtaining an improved understanding the various roles of PAK6 in the cell, its substrates and autoregulation, its importance in disease and its potential targeted inhibition. Regulation of type II PAKs was poorly understood until recently. Unlike many protein kinases where phosphorylation at conserved sites within the so-called ��activation loop�� is a critical step towards full activity, the type II PAKs are constitutively phosphorylated in the cell and not directly regulated by interaction with small GTPases, which are instead important for type II PAK relocalization. We, and others, identified an autoinhibitory sequence within the N-terminal region of PAK4 and showed by structural and biochemical analysis that this region contains a pseudosubstrate sequence centered around residue P52. Based on this work, we hypothesized that this highly conserved N-terminal region could autoinhibit each of the type II PAKs. ATP-competitive small molecule inhibitors of the type II PAKs could be useful as cancer therapeutics. The small molecule PF-3758309 was designed as a PAK4-specific inhibitor, but displays in vitro activity against each of the type II PAKs and also PAK1. Though effective in mouse models of cancer, it failed in human clinical trials. Sunitinib is a potent ATPcompetitive multi-kinase inhibitor that is indicated for treatment of renal cell carcinoma, imatinib-resistant gastrointestinal stromal tumors, advanced pancreatic neuroendocrine tumors and other tumor types. A crystal structure is available for PAK4 with PF-3758309 but none are available for a PAK family member in complex with sunitinib. In the current study we ask whether downstream substrate specificity is conserved among the type II PAKs, whether a cancerassociated mutation that occurs in the type II PAK autoinhibitory region can activate PAK6, and whether co-crystallography might aid drug discovery for type II PAKs. By peptide array profiling we show that PAK6 has a similar substrate specificity to that previously observed for PAK4 and PAK5, implying that PAK6 may have additional substrates that overlap with other type II PAKs. We show that PAK6 kinase activity is regulated by its Nterminal pseudosubstrate in vitro and that a melanoma-associated mutation, P52L, in the pseudosubstrate sequence displays reduced inhibition. We go on to determine crystal structures of PAK6 kinase domain in complex with two ATP-competitive small molecule inhibitors, PF-3758309 and sunitinib. This study therefore provides molecular level details that may aid in the development of isotype specific inhibitors for the type II PAKs.
alternate mechanisms of activation and regulation have recently been discovered. The type II PAKs are important for signaling LY294002 cascades that regulate cell survival, neurite outgrowth and formation of filipodia. PAK6 is expressed in prostate, testis, thyroid, placenta and neural tissues and is found in both cytoplasmic and nuclear fractions of prostate cells. Androgen receptor is reported to be a downstream target of PAK6, and PAK6 can regulate gene transcription by androgen receptor via a GTPase-independent mechanism possibly related to control of its degradation by the MDM2 E3 ubiquitin ligase. Global deletion of Pak6 in mice results in increased weight and decreased aggression, possibly explained by its role in androgen receptor signaling. In addition, mice with combined deletion of Pak5 and Pak6 show deficits in locomotion, learning and memory not associated with single deletions of either gene, suggesting functional redundancy between the two PAKs. While neuronal substrates specific to PAK6 have not been identified, PACSIN1, an FBAR protein involved in synaptic vesicle recycling, is phosphorylated redundantly by PAK4, PAK5 and PAK6 in vivo PAK6 is overexpressed in prostate cancer, and its targeted inhibition could potentially decrease growth of prostate tumors or sensitize prostate cancer cells to radiotherapy. PAK6 has also been found to acquire somatic mutations in other solid tumors, including mutation of residue Pro52 to leucine in two independent melanomas. Consequently there is increasing interest in obtaining an improved understanding the various roles of PAK6 in the cell, its substrates and autoregulation, its importance in disease and its potential targeted inhibition. Regulation of type II PAKs was poorly understood until recently. Unlike many protein kinases where phosphorylation at conserved sites within the so-called ��activation loop�� is a critical step towards full activity, the type II PAKs are constitutively phosphorylated in the cell and not directly regulated by interaction with small GTPases, which are instead important for type II PAK relocalization. We, and others, identified an autoinhibitory sequence within the N-terminal region of PAK4 and showed by structural and biochemical analysis that this region contains a pseudosubstrate sequence centered around residue P52. Based on this work, we hypothesized that this highly conserved N-terminal region could autoinhibit each of the type II PAKs. ATP-competitive small molecule inhibitors of the type II PAKs could be useful as cancer therapeutics. The small molecule PF-3758309 was designed as a PAK4-specific inhibitor, but displays in vitro activity against each of the type II PAKs and also PAK1. Though effective in mouse models of cancer, it failed in human clinical trials. Sunitinib is a potent ATPcompetitive multi-kinase inhibitor that is indicated for treatment of renal cell carcinoma, imatinib-resistant gastrointestinal stromal tumors, advanced pancreatic neuroendocrine tumors and other tumor types. A crystal structure is available for PAK4 with PF-3758309 but none are available for a PAK family member in complex with sunitinib. In the current study we ask whether downstream substrate specificity is conserved among the type II PAKs, whether a cancerassociated mutation that occurs in the type II PAK autoinhibitory region can activate PAK6, and whether co-crystallography might aid drug discovery for type II PAKs. By peptide array profiling we show that PAK6 has a similar substrate specificity to that previously observed for PAK4 and PAK5, implying that PAK6 may have additional substrates that overlap with other type II PAKs. We show that PAK6 kinase activity is regulated by its Nterminal pseudosubstrate in vitro and that a melanoma-associated mutation, P52L, in the pseudosubstrate sequence displays reduced inhibition. We go on to determine crystal structures of PAK6 kinase domain in complex with two ATP-competitive small molecule inhibitors, PF-3758309 and sunitinib. This study therefore provides molecular level details that may aid in the development of isotype specific inhibitors for the type II PAKs.
In binding models only the propyl group is within the A-site while the naphthalene backbone is mostly outside
Three systems, LDHA:0SN, LDHA:2B4, and LDHA:NHIS, could hold the mobile loop in the closed conformation. Additionally, the mobile loop displayed larger fluctuations in the open conformation than in the closed conformation, which is probably caused by a much larger conformational space available for the loop open state. It follows that bringing the mobile loop to the closed conformation causes an entropic penalty. This could partially explain the comparable binding affinities of 0SN and 1E4, even though 0SN possesses more polar interactions. Similarly, the ionic interactions with Arg111 were shown to significantly reduce the mobility of 1E4 and surrounding A-site residues, including Arg111; the incurred entropic penalty would offset the enthalpy gain from such strong ionic interactions. Since Arg111 is largely exposed to bulk solvent, polar water molecules can also compete with the inhibitor in interacting with Arg111. Notably, similar ionic interactions in the LDHA:1E7 complex appeared to be unstable, suggesting little free energy gain from this interaction. No significant correlation between the dynamics of ligand binding, as revealed by RMSF values of binding site residues and ligands as well as the percentage existence of polar interactions, and experimental binding affinities was found. For example, the binding of 1E4 incurred much larger fluctuations with smaller percentage existence of polar interactions than that of 0SN, but their experimental binding affinities are  roughly the same, with 1E4 being slightly higher. The same phenomenon was observed for A-site binders 1E7 and AJ1. Likewise, the number of strong polar interactions or contactsdoes not predict the strength of binding. Hence, conventional MD simulations appear to be incapable of discriminating LDHA inhibitors of different binding strengths. To resolve this issue, we resorted to steered MD simulations, which can qualitatively discern inhibitors of largely different binding affinities. Steered MD simulations have demonstrated the effects of different initial conformations of the mobile loopand different sites of bindingon the difficulty of pulling. Considering these effects, our pulling results correlated well with experimental binding affinities and were able to distinguish inhibitors with a small 4 kJ mol21 DGdissoc difference, despite their different dynamics and modes of binding. Although DPMF values, calculated from exponential averages of SP600125 JNK inhibitor non-equilibrium work, largely depend on rarely sampled trajectories with small dissipated work, the work and peak force were able to qualitatively discriminate inhibitors of the same binding site and initial loop conformation. Other computational approaches such as umbrella BU 4061T sampling can yield a better estimate of free binding energy.Nevertheless, steered MD simulations provide a more convenient set-up with much less computational cost for ranking inhibitors with respect to relative binding affinities. Our steered MD simulations also suggest that NHI is more likely to bind in the A-site by comparison of relative difficulties in pulling, even though NHI binding models in both the A-site and the S-site, generated from conventional MD simulations, can explain its experimental structure-activity relationships.After all, NHI behaved differently in the S-site from other inhibitors that have only one carboxylate group within the S-site, in that NHI could hold the mobile loop closed by interacting with Arg105 for most of the time while others could not. The binding of NHI at the A-site also agrees with preliminary NMR and crystallographic data.On the other hand, our attempts to obtain possible binding modes of FX11 were unsuccessful.
roughly the same, with 1E4 being slightly higher. The same phenomenon was observed for A-site binders 1E7 and AJ1. Likewise, the number of strong polar interactions or contactsdoes not predict the strength of binding. Hence, conventional MD simulations appear to be incapable of discriminating LDHA inhibitors of different binding strengths. To resolve this issue, we resorted to steered MD simulations, which can qualitatively discern inhibitors of largely different binding affinities. Steered MD simulations have demonstrated the effects of different initial conformations of the mobile loopand different sites of bindingon the difficulty of pulling. Considering these effects, our pulling results correlated well with experimental binding affinities and were able to distinguish inhibitors with a small 4 kJ mol21 DGdissoc difference, despite their different dynamics and modes of binding. Although DPMF values, calculated from exponential averages of SP600125 JNK inhibitor non-equilibrium work, largely depend on rarely sampled trajectories with small dissipated work, the work and peak force were able to qualitatively discriminate inhibitors of the same binding site and initial loop conformation. Other computational approaches such as umbrella BU 4061T sampling can yield a better estimate of free binding energy.Nevertheless, steered MD simulations provide a more convenient set-up with much less computational cost for ranking inhibitors with respect to relative binding affinities. Our steered MD simulations also suggest that NHI is more likely to bind in the A-site by comparison of relative difficulties in pulling, even though NHI binding models in both the A-site and the S-site, generated from conventional MD simulations, can explain its experimental structure-activity relationships.After all, NHI behaved differently in the S-site from other inhibitors that have only one carboxylate group within the S-site, in that NHI could hold the mobile loop closed by interacting with Arg105 for most of the time while others could not. The binding of NHI at the A-site also agrees with preliminary NMR and crystallographic data.On the other hand, our attempts to obtain possible binding modes of FX11 were unsuccessful.
Several indole derivatives compoundsthat mediate direct and selective interactions with members of the serine-arginine
Certain IDC have been proven to be potent inhibitors of HIV-1 replication in cell culture through a selective action on exonic splicing enhancers -dependent activity of individual SR proteins. One such molecule, IDC16 has been shown to interfere with the SF2/ASF SR protein and production of HIV regulatory proteins and to compromise assembly of infectious particles. However, no evaluation of IDC on retrovirus-mediated pathogenesis has yet been documented. Here, we have taken advantage of the F-MLV induced pathogenesis model in newborn mice to evaluate the efficiency of this new class of molecules at different stages of retrovirus infection and disease. We show now that different IDC differentially inhibit HIV-1 and MLV, most likely reflecting distinct requirement for cellular splicing factors. Thus, we found that IDC13 and IDC78, but not IDC16, prevented F-MLV replication both ex vivo and in vivo, by selectively altering single-splicing of the retroviral genome. Furthermore, we describe two IDC that also proved to be very efficient at protecting mice from MLV-induced pathogenesis by inhibiting early viral replication. In order to better understand the molecular mechanisms underlying the specific inhibition of MLV replication by some of the IDC, we analyzed the viral RNA content of infected cells. Dunni cells were infected with F-MLV in the presence of different IDC and total RNA was extracted and used as template for RTPCR. We used two different sets of oligonucleotide primers that allowed us to discriminate between spliced and unspliced viral RNAs. As an internal control, RT-PCR was performed on mRNA from the gadph house-keeping gene. Compared to untreated cells, accumulation of the PCR product corresponding to the spliced F-MLV RNA dramatically decreased upon treatment with IDC13 and IDC78, while accumulation of the gapdh product did not decrease. Neither IDC16, mentioned above, nor IDC217, a compound that had no MK-0683 molecular weight effect on all splicing substrates tested, had detectable impact 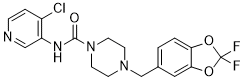 on F-MLV splicing. Altogether, these results indicated that inhibition of F-MLV replication by certain IDC appeared directly associated with their ability to specifically inhibit viral RNA splicing, an event required for expression of the viral Env glycoprotein. However, we observed that the significant decrease of spliced product observed after IDC13 and IDC78 treatment was not compensated by a corresponding increase of unspliced FMLV RNA. Instead, we noted that IDC13, IDC78 and also IDC16 affected, albeit to a lesser extent for the latter, accumulation of unspliced viral RNA. These results suggested that IDC13 and IDC78 inhibited F-MLV replication by altering viral RNA splicing, but that other pathway governing RNA accumulation, such as transcriptional levels, RNA trafficking and/or RNA stability, could also be altered. Furthermore, we observed that there was a correlation between a lower plasmatic viremia and increased latency of disease in CPI-613 IDC-treated mice. Altogether, this indicated that lower virulence of F-MLV, observed in IDC-treated mice, was likely due to inhibition of virus replication during the early phase of the disease. In order to further assess whether resistance to F-MLV-induced erythroleukemia in IDC-treated mice was indeed due to inhibition of earlyvirus dissemination, and nottoa toxic effect leadingtoreduction of target cells, we measured the direct effect of these compounds on erythroid differentiation in vivo. Newborn mice were injected with compounds IDC13, IDC78 or IDC217and followed both for hematocrits between 16 to 21 days of age and for spleen enlargement as an indication of compensatory splenic activity.
on F-MLV splicing. Altogether, these results indicated that inhibition of F-MLV replication by certain IDC appeared directly associated with their ability to specifically inhibit viral RNA splicing, an event required for expression of the viral Env glycoprotein. However, we observed that the significant decrease of spliced product observed after IDC13 and IDC78 treatment was not compensated by a corresponding increase of unspliced FMLV RNA. Instead, we noted that IDC13, IDC78 and also IDC16 affected, albeit to a lesser extent for the latter, accumulation of unspliced viral RNA. These results suggested that IDC13 and IDC78 inhibited F-MLV replication by altering viral RNA splicing, but that other pathway governing RNA accumulation, such as transcriptional levels, RNA trafficking and/or RNA stability, could also be altered. Furthermore, we observed that there was a correlation between a lower plasmatic viremia and increased latency of disease in CPI-613 IDC-treated mice. Altogether, this indicated that lower virulence of F-MLV, observed in IDC-treated mice, was likely due to inhibition of virus replication during the early phase of the disease. In order to further assess whether resistance to F-MLV-induced erythroleukemia in IDC-treated mice was indeed due to inhibition of earlyvirus dissemination, and nottoa toxic effect leadingtoreduction of target cells, we measured the direct effect of these compounds on erythroid differentiation in vivo. Newborn mice were injected with compounds IDC13, IDC78 or IDC217and followed both for hematocrits between 16 to 21 days of age and for spleen enlargement as an indication of compensatory splenic activity.
Required to improve the outcome of CML patients carrying the T315I mutation
Ponatinib, also known as AP24534, is an oral, multi-targeted TKI. Olaparib distributor Ponatinib is effective at nanomolar levels against T315I and other point mutations. This TKI has been investigated in a pivotal phase 2 clinical trial in patients with resistant or intolerant CML and Ph-positive acute lymphoblastic leukemia. Histone acetyltransferases and histone deacetylasesfunction antagonistically to control histone acetylation. HDACs regulate chromatin remodeling and are crucial in the epigenetic regulation of various genes. Abnormal activity or expression of HDACs has been found in a broad range of tumor types. An HDAC inhibitorblocks the activity of specific HDACs. Preclinical data suggesta role for HDACi as apotential new treatment in several tumor types, including hematological malignancies. In this study, we investigated ponatinib activity against Phpositive leukemia cells carrying the T315I mutation. We also examined the efficacy of HDACi vorinostat in combination with ponatinib in various cell lines. This study also aimed to explore the molecular mechanism of ponatinib resistance by using BCR-ABLexpressing cell lines with point mutations. Furthermore, cotreatment with ponatinib and vorinostat suppressed growth in ABL TKI ponatinib-resistant clones. Therefore, we examined the activity of an HDACi against T315I mutant cells. We found that another HDACi, vorinostat, was effective against Ba/F3 T315I cells. We examined whether treatment with ponatinib and vorinostat induced cell death in T315I mutant cells. We found that treatment with ponatinib and vorinostat significantly inhibited T315I mutant cell growth compared to treatment with single drugs. Twoway ANOVA analysis Vismodegib Hedgehog inhibitor revealed that the combined effects of the drugs was synergistic. We also investigated intracellular signaling and found that after BCR-ABL phosphorylation, Crk-L activity reduced, PARP cleaved, and cH2A.X phosphorylation increased after ponatinib and vorinostat treatment. We next examined ponatinib and vorinostat co-treatment in primary Ph+ leukemia samples. The clinical data for the CML- and Ph-positive leukemia patients are documented. Analysis of whole blood cells with BCR-ABL fluorescence in situ hybridization showed that.95% of cells were BCR-ABL positive. Ponatinib is effective against T315I mutant cells that are resistant to imatinib and second-generation ABL TKIs nilotinib and dasatinib. O��Hare and colleagues reported that treatment with 40 nM ponatinib did not yield any BCR-ABL mutant cells. We confirmed that ponatinib was effective against BCR-ABL wild-type and T315I mutant cells at low concentrations by cell proliferation 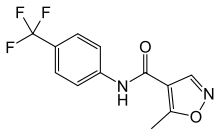 and immunoblot assays. An important finding in this study was that combined treatment with ponatinib and vorinostat showed antiproliferative effects in vitro and exhibited antitumor activity in vivo. Using the Ba/F3 T315I xenograft model, ponatinib or vorinostatshowed similar reduction in tumor size. We demonstrated the tumor volumes in mice treated with both ponatinib and vorinostat were significantly reduced compared to those treated with each drug alone. Immunohistochemical analysis revealed that the expression of the proliferation marker Ki67 reduced and TUNEL-positive cells increased in ponatinib and vorinostat-treated mice. These results suggest that this combination was effective against T315I mutation in vivo. Overall, the results indicate that a greater level of efficacy was achieved with combined treatment with ponatinib and vorinostat. Multiple preclinical studies and clinical data support the use of HDACis in combination with other drugs for the treatment of various cancers, including leukemia. Some HDACis, including vorinostat and romidepsin, have been approved for use against cutaneous T-cell lymphoma.
and immunoblot assays. An important finding in this study was that combined treatment with ponatinib and vorinostat showed antiproliferative effects in vitro and exhibited antitumor activity in vivo. Using the Ba/F3 T315I xenograft model, ponatinib or vorinostatshowed similar reduction in tumor size. We demonstrated the tumor volumes in mice treated with both ponatinib and vorinostat were significantly reduced compared to those treated with each drug alone. Immunohistochemical analysis revealed that the expression of the proliferation marker Ki67 reduced and TUNEL-positive cells increased in ponatinib and vorinostat-treated mice. These results suggest that this combination was effective against T315I mutation in vivo. Overall, the results indicate that a greater level of efficacy was achieved with combined treatment with ponatinib and vorinostat. Multiple preclinical studies and clinical data support the use of HDACis in combination with other drugs for the treatment of various cancers, including leukemia. Some HDACis, including vorinostat and romidepsin, have been approved for use against cutaneous T-cell lymphoma.