Given its intriguing position at the crossroads of these two processes, we were interested in investigating the expression and regulation of SelS. In this study we show that only one of the human SelS mRNA variants can encode a Folinic acid calcium salt pentahydrate selenoprotein of 189 amino acids. The other transcript encodes a truncated protein of 187 amino acids that lacks selenocysteine. Additionally, elements in the 39UTR of the selenoprotein-encoding mRNA positively and negatively influence Sec insertion into SelS, and provide another mechanism to regulate the production of these two protein isoforms. The ability of 39UTR elements to influence the incorporation of Sec underscores the importance of context when examining functional RNA elements such as the SECIS. We also show that in addition to being an ER-resident protein, the subcellular localization of endogenous SelS includes enrichment at perinuclear speckles adjacent to the Golgi, which was previously unknown. The color of the base indicates the likelihood of its involvement in a base pairing interaction. The probability scale runs from blue to red. In addition, positions where compensatory mutations occur in the sequence set are indicated on the structure with black circles around the nucleotides. SL1 displays a high probability of forming a stem-loop structure, as the majority of the structure registers in the red range. The only exception is the base pair at the top of the stem, which likely reflects a tolerance for the helix to breathe at this position. For SL2, the 50 nucleotides immediately downstream from the SECIS element were used to generate the alignment. The location of the SECIS in each sequence was defined using SECISearch. Figure 5A shows the LOUREIRIN-B RNAalifold structure annotated sequence alignment for this region. This region of the SelS 39UTR retains its AU-rich character across the sequence set but it is more difficult to discover sequence covariance in the region, particularly with the inclusion of non-mammalian species. Despite the sequence noise, Figure 5B shows the high-probability formation of a stem-loop structure in this region. The likelihood of the base pair interactions across the predicted stem is reinforced by the detection of compensatory mutations 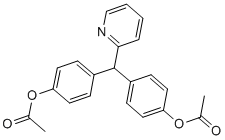 for each position, as indicated by black circles around the nucleotides involved. As the set of sequences is heavily weighted to mammals, we also conducted a pairwise analysis using the Ciona and Xenopus sequences in the combined locARNA/RNAalifold analysis. This analysis also predicts the formation of a stem-loop of similar size and length. Thus, the ability to form a stem-loop structure downstream of the SECIS element is not restricted to mammals. While the dampening effect of the 39UTR on the SelS SECIS is from downstream sequences, we were still interested in examining the upstream element SL1 for possible effects on Sec insertion. One could envision SL1 exerting a positive effect on Sec insertion by promoting ribosome pausing during translation. Conversely, SL1 could have a negative impact on selenoprotein synthesis by preventing the recoding machinery from accessing the UGA codon. The relative distance between this stem-loop and the UGA codon is very different in the endogenous and heterologous contexts. In its native context, SL1 is 9 nucleotides downstream of the UGA codon, whereas in the luciferase reporter there are several hundred nucleotides between them.
for each position, as indicated by black circles around the nucleotides involved. As the set of sequences is heavily weighted to mammals, we also conducted a pairwise analysis using the Ciona and Xenopus sequences in the combined locARNA/RNAalifold analysis. This analysis also predicts the formation of a stem-loop of similar size and length. Thus, the ability to form a stem-loop structure downstream of the SECIS element is not restricted to mammals. While the dampening effect of the 39UTR on the SelS SECIS is from downstream sequences, we were still interested in examining the upstream element SL1 for possible effects on Sec insertion. One could envision SL1 exerting a positive effect on Sec insertion by promoting ribosome pausing during translation. Conversely, SL1 could have a negative impact on selenoprotein synthesis by preventing the recoding machinery from accessing the UGA codon. The relative distance between this stem-loop and the UGA codon is very different in the endogenous and heterologous contexts. In its native context, SL1 is 9 nucleotides downstream of the UGA codon, whereas in the luciferase reporter there are several hundred nucleotides between them.