Each set had clinical follow-up information in form of censored time to event data, the event being either “overall survival” or “relapse-free survival” or both. The goal was to extract a gene signature from a training set that can be used to predict disease outcome for patients in the testing set. The gene signature we used consisted of a set of genes plus corresponding Cox coefficients derived by univariate fitting of the expression values to the survival data on the training set. Gene signatures were built from the individual or merged data sets. The accuracy and robustness of prediction were evaluated by 10-fold cross-validation. Reproducibility as defined above was analyzed by training a signature from one or several complete data sets and testing its performance on complete independent validation sets. Data sets were merged in their original numerical representation using two different data integration methods: ComBat and Z-score normalization. Two signature performance measures were computed in each experiment: time dependent Receiver-Operator Characteristic Area Under the Curve and the hazard ratio of the Echinatin predicted 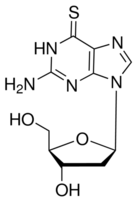 risk scores relative to the survival data in the testing set. Note that the latter required stratification of the testing set patients in a high and low risk groups. In total, we analyzed 1324 breast cancer samples from public data sets generated with three microarray technologies. To the best of our knowledge, this study is the largest one evaluating the potential benefits of data merging in a quantitative OS/RFS patients risk prediction framework. They also reveal the limited usefulness of the data intermingling test, which in this case provides a misleading Pimozide picture of the variance retained after data integration. Noting that the gene signatures built from subsets of GSE4335 or Vijver showed higher prediction accuracies in cross-validation than the gene signature built from the merged data set, we investigated how the performance could possibly be improved by selective data integration. To assess the reproducibility of the gene signatures’performance derived from the merged data sets, the prediction accuracy was evaluated in a leave-one-data set-out manner. In each step, one complete source data set was set aside as testing set while the predictor was built from the merged remaining sets.
risk scores relative to the survival data in the testing set. Note that the latter required stratification of the testing set patients in a high and low risk groups. In total, we analyzed 1324 breast cancer samples from public data sets generated with three microarray technologies. To the best of our knowledge, this study is the largest one evaluating the potential benefits of data merging in a quantitative OS/RFS patients risk prediction framework. They also reveal the limited usefulness of the data intermingling test, which in this case provides a misleading Pimozide picture of the variance retained after data integration. Noting that the gene signatures built from subsets of GSE4335 or Vijver showed higher prediction accuracies in cross-validation than the gene signature built from the merged data set, we investigated how the performance could possibly be improved by selective data integration. To assess the reproducibility of the gene signatures’performance derived from the merged data sets, the prediction accuracy was evaluated in a leave-one-data set-out manner. In each step, one complete source data set was set aside as testing set while the predictor was built from the merged remaining sets.